NutriScoPe – Automated diagnosis of malnutrition (nutritional scoring) in inpatients
Project Period: 11/2024 – 10/2025
Funding: TMF e.V.
Project Description
NutriScoPe is a retrospective study examining the occurrence of malnutrition in patients across various hospital departments and assessing the impact of systematic nutritional assessment and therapy on treatment outcomes.
Malnutrition affects up to 20–30% of hospitalized patients and is associated with longer hospital stays, higher costs, and increased mortality—particularly in patients with malignant diseases or chronic organ failure. To address these risks, a dedicated nutrition team was established at the University of Leipzig Medical Center (UKL) to provide targeted support to patients during their inpatient stay.
At UKL, patients at increased risk of malnutrition are identified using a standardized procedure called Nutritional Risk Screening (NRS). Upon hospital admission, patients undergo an initial prescreening, and if abnormalities are detected, a main screening follows. Patients diagnosed with malnutrition then receive targeted nutritional therapy from the nutrition team. In contrast, no systematic NRS screening is currently implemented at Jena University Hospital (UKJ).
The study utilizes medical documentation data from both hospitals to analyze the prevalence of malnutrition, its distribution across different clinical departments, and the effects of automated Nutritional Risk Screening on treatment success, length of hospital stay, and complications. Additionally, associations with age, sex, and primary diagnoses are examined to derive targeted measures for improving patient care.
Team Members
Project Partners
Results
The data analysis showed that not all patients with a positive prescreening underwent a subsequent main screening at UKL, representing a deviation from the NRS protocol. In clinical practice, the main screening was performed predominantly in patients with more severe disease courses.
Since the prescreening is based on self-assessment at hospital admission, some bias was expected. Notable main screenings were particularly observed in the field of visceral surgery, often in patients with malignant comorbidities.
A higher NRS score in the main screening was strongly correlated with poorer clinical outcomes. These patients showed increased mortality, longer hospital stays, and higher readmission rates.
The following figure illustrates the key results of the analysis to date (as of September 2025):
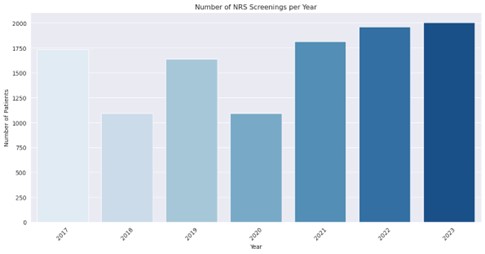
Figure 1: Number of patients who underwent nutritional risk screening (NRS screening) at UKL between 2017 and 2023. Illustration by the author.
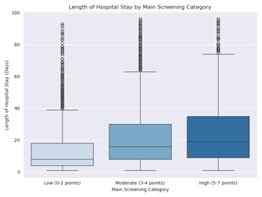
Figure 2: Length of inpatient stay depending on NRS score grouping
in the main screening
| 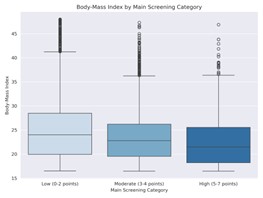
Figure 3: Body mass index depending
on NRS score grouping
in the main screening
|
Publications
- Pirlich, M., Schütz, T., Norman, K., Gastell, S., Lübke, H. J., Bischoff, S. C., Bolder, U., Frieling, T., Güldenzoph, H., Hahn, K., Jauch, K. W., Schindler, K., Stein, J., Volkert, D., Weimann, A., Werner, H., Wolf, C., Zürcher, G., Bauer, P., & Lochs, H. (2006). The German hospital malnutrition study. Clinical nutrition (Edinburgh, Scotland), 25(4), 563–572. Read paper
- Schuetz, P., Fehr, R., Baechli, V., Geiser, M., Deiss, M., Gomes, F., Kutz, A., Tribolet, P., Bregenzer, T., Braun, N., Hoess, C., Pavlicek, V., Schmid, S., Bilz, S., Sigrist, S., Brändle, M., Benz, C., Henzen, C., Mattmann, S., Thomann, R., … Mueller, B. (2019). Individualised nutritional support in medical inpatients at nutritional risk: a randomised clinical trial. Lancet (London, England), 393(10188), 2312–2321. Read paper