Physiological relevance of msGPCRs as mediators between microbiota and the human host
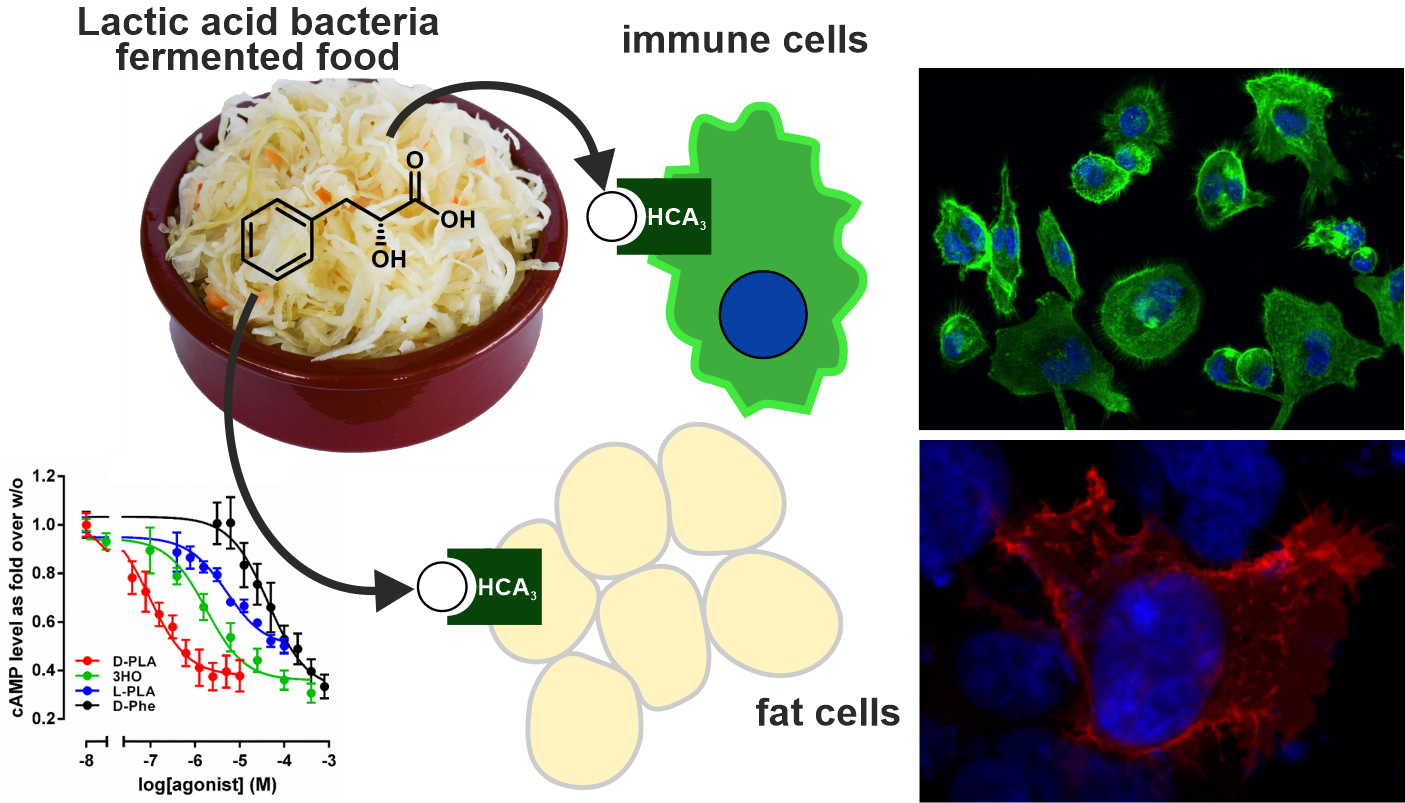
We apply a broad interdisciplinary approach that combines evolutionary, functional, pharmacological, immunological and pharmacokinetic methods to analyze msGPCRs. We successfully applied this strategy to unravel the role of hydroxycarboxylic acid receptor 3 (HCA3), a hominid-specific receptor expressed in immune cells and adipocytes. We discovered that lactic acid bacteria (LAB) fermented food-derived metabolites are highly potent agonists at this receptor. In an evolutionary context, this suggests that the availability of a new food repertoire under changed ecological conditions triggered the fixation of HCA3, which took over new functions in hominids. Moreover, we could show that HCA3 activation results in anti-inflammatory responses in immune cells and anti-lipolytic effects in adipocytes.
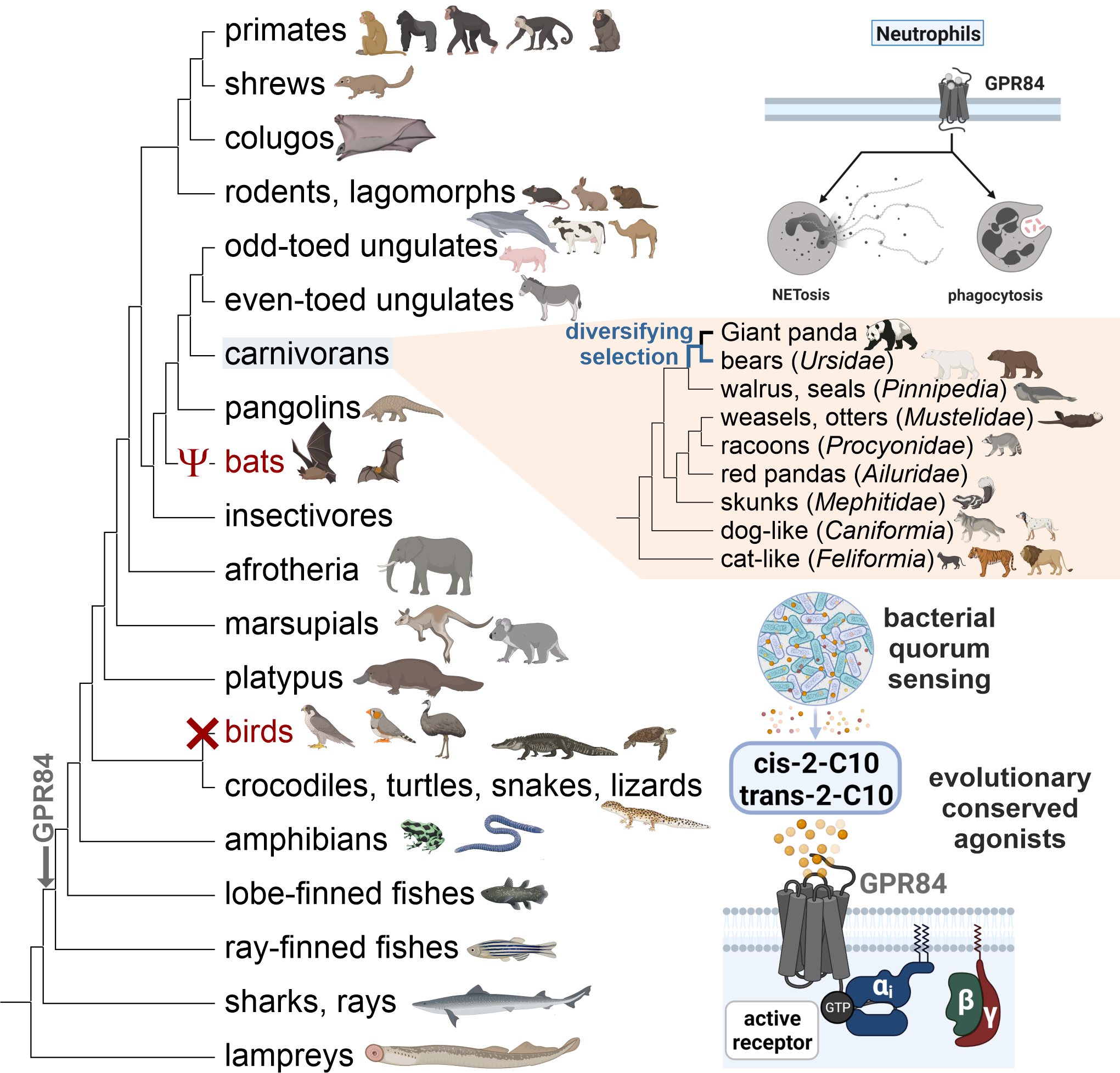
The relevance of microbial colonization for healthy functional organisms increasingly gains attention. Since the microbiome is species-specific and dependent on habitat and diet, we believe that an evolutionary approach to understanding the relevance of msGPCRs in this context is highly adequate.

![]()
![]()
Pharmacology, signal transduction, trafficking and subcellular distribution of msGPCRs
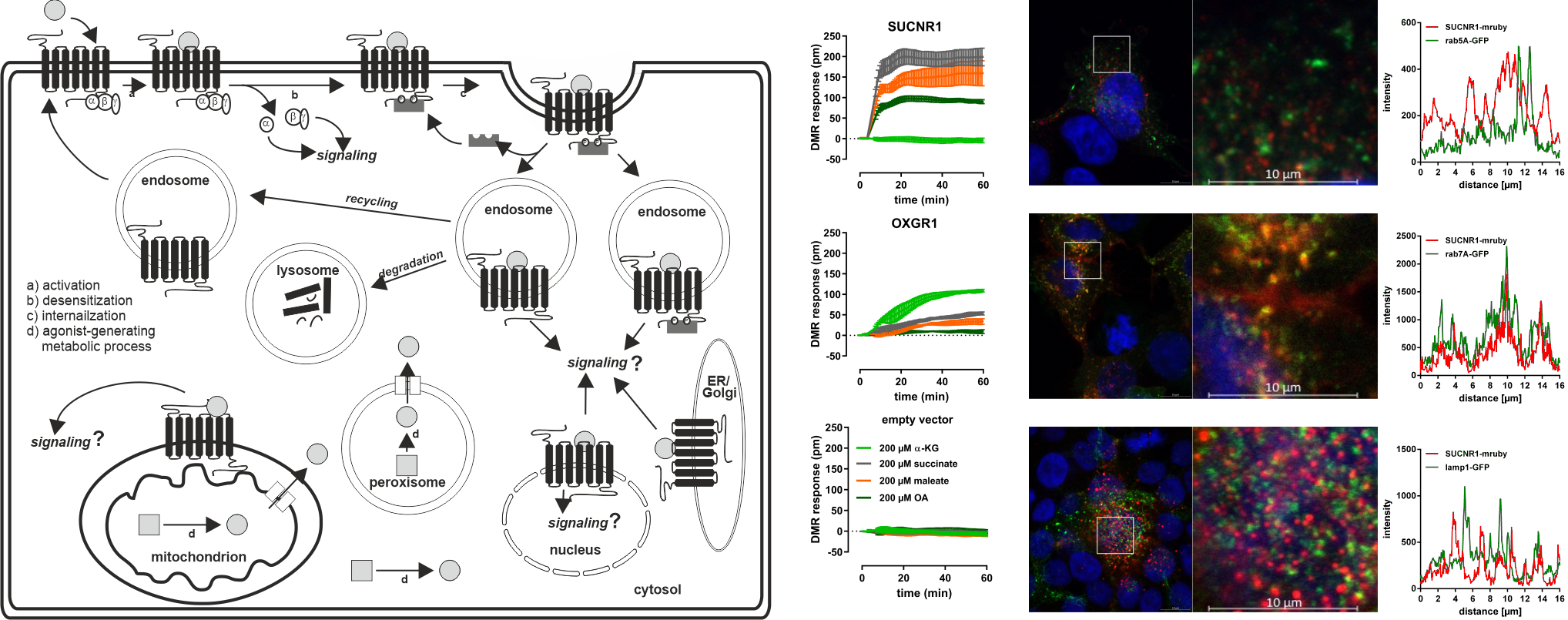
It is very important to better understand the pharmacology, trafficking and signal transduction of these receptors since some msGPCRs are activated by metabolites that occur mainly intracellularly in effective concentrations. It is well-accepted that GPCRs signal from the plasma membrane and detect extracellular ligands, but there is accumulating evidence for GPCR signaling from intracellular membranes, such as endosomes and mitochondria. Especially under certain pathological conditions, some metabolites are increasingly released intracellularly from e.g. mitochondria.
Our group systematically aims to test the hypothesis that some of these msGPCRs can reside and signal from intracellular compartments. With these analyses, we aim to increase the knowledge of the molecular mechanisms and cellular metabolic adaptations mediated by msGPCRs and extend our understanding of the subcellular distribution, signaling, and pharmacology of GPCRs activated by energy metabolites.
The role of msGPCRs in cancer cell metabolism
Cancer cell metabolism is rendered to support rapid proliferation and is characterized by certain metabolic features, including altered glucose, fatty acid and glutamine metabolism, when compared to normal proliferating cells. Up until now, the role of msGPCRs in regulating cancer cell metabolism is insufficiently understood.
We aim to understand, which msGPCR-activated signaling pathways influence the metabolism of cancer cells in which way. We analyze viability, proliferation, and cytotoxicity in combination with biochemical and pharmacological analyses to understand the link between msGPCRs and cancer cell metabolism. Further, we apply metabolomics analyses using global targeted and untargeted Liquid Chromatography Mass Spectrometry (LC-MS) profiles, the seahorse analyzer that determines oxygen consumption rate and extracellular acidification rate as well as FRET metabolite sensors. We analyze 2D cancer cell lines and their derived 3D tumor spheroids.